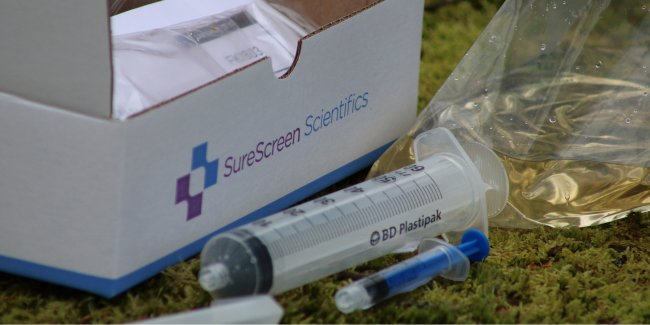
eDNA Filtration Kit sample collection guide
- Articles
- Sample Collection Guidance


Step 1
Identify where 20 sub-samples will be taken from the river or pond perimeter.
The location of these should be spaced as evenly as possible around the site. In ponds, samples should be taken from locations around the entire pond perimeter, where accessibility permits. In rivers, samples should be taken against the flow of the stream, working upstream in a diagonal pattern where possible. This will ensure that any disturbed sediment is not collected, should it be necessary for the collector to enter the watercourse.
Step 2
Wearing gloves, open the sterile Whirl-Pak bag and collect 20 ladles of water from the 20 sub-sites.
The water sample should be taken from the middle of the water column (at least 10cm from bottom where possible). Where possible, avoid any disruption of sediment as this can both quickly clog the filter and introduce ancient DNA into the sample. In larger sites it may be necessary to use a telescopic pole.
Once collected close the bag securely and shake to mix the water sample.
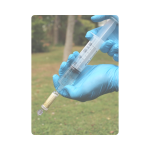
Step 3
Using the large syringe, take 50ml of sample from the Whirl-Pak bag.
Attach the syringe using a half twist action to the filter unit. The syringe will only fit to one end of the filter unit. Note, twisting too far can damage the luer lock connection on the filter.
Apply pressure to the syringe until all liquid has passed into and through the filter.
Remove the syringe from the filter and repeat the process until:
The more liquid passed through the filter unit, the more reliable results will be, however, be careful not to push too hard as the filter casing can crack under extreme pressure. Record the volume of liquid which has been filtered on the sample collection form.

Step 4
Empty the syringe and fill with air, re-attach to the filter and push air through the filter unit until it is completely free of water.
Step 5
Screw the spare red cap tightly onto the thick end of the filter unit.
Place the filter unit to one side.

Step 6
Remove the cap from the small syringe and store to one side.
An excess of preservative solution is provided.
It is important to add preservative solution into the filter unit to prevent sample degradation during transport to the laboratory.
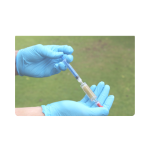
Step 7
Attach the syringe to the open end of the filter unit.
Slowly apply light pressure until the filter casing is filled with preservative solution.
The preservative solution allows for the filter to be stored at room temperature before analysis. In the absence of a preservative, filters must be frozen immediately and returned to the lab on ice.
Step 8
Finally, screw the red cap on to the other end of the filter casing.
Ensure that both caps are secured tightly to avoid leakage of preservative solution during transport to the laboratory.
Place the sample into the 50ml tube provided and return to laboratory.
Samples can be stored at room temperature for up to 2 weeks, 4 weeks if chilled.


Yes. Each sample kit can be used to detect up to 4 eDNA target species. For more than 4 species you will need to use additional sampling kit. Please contact us for an up-to-date list of the target species we can detect, to ensure that your species are on our list.
This depends on the type and size of the study site, and the question that you are trying to answer. For a small, isolated site such as a pond, small lake or isolated river site one sample should be sufficient. For larger river systems and reservoirs bigger than 1-hectare, multiple samples should be taken from multiple sites to ensure the site is best represented by the sampling strategy. If you are struggling to determine how many samples to collect for a study site, then get in touch with us and we will be able to advise.
If you require a high level of scientific accuracy, for example, when screening sites for the presence of disease or to triple-check an ark site is free of an invasive species, we recommend that you collect multiple replicate samples (over a period of time/on different dates/using different sub-sample sites) from each site to ensure consistency and reliability in results.
Anyone can sample! A license is only required if you are conducting a protected species survey, in addition to the eDNA survey. If you are just simply taking a water sample and not intending to search for the target species (such as white-clawed crayfish) or disturb their habitat then you do not require a licence.
The assays used in the laboratory for the detection of the target species have by design been developed as species specific. This means that they will only detect and amplify DNA of the target species. Thorough testing has been conducted to ensure that this is the case and that each of our eDNA assays do not cross-amplify/identify any other non-target species.
The filters are designed to process up to 1 litre of fluid. The more liquid passed through the filter unit, the more reliable results will be, however, be careful not to push too hard as the filter casing can crack under extreme pressure. In many cases, due to turbidity and sediment load it may not always be possible to filter a high volume. We recommend filtering up to 500ml (minimum required volume = 150ml). Filter until you reach either 500ml/the filter becomes clogged/you are no longer able to push any liquid through, using a reasonable amount of pressure. Do not push beyond the point where it takes more than 30 seconds to filter 50ml. Should the filter unit crack there is an increased chance of DNA degradation before analysis.
All of our filter based eDNA kits have an expiry date of 1 year from manufacture. If the kit is kept in good conditions (cool, dark) it can in most cases be used after a few months beyond this date.
It is best to coincide sample collection with favourable sampling conditions which are based upon seasonal weather and seasonal changes in your target species’ activity levels. For example: We recommend that crayfish samples should be collected between mid-April and late-October to coincide with when crayfish are most active (and hence releasing most eDNA). Samples can be taken outside of this window but may not provide highly reliable or repeatable results.
Further to this, the sample should be collected when the pond/river/stream is relatively calm, with little turbidity. Try to avoid sample collection from murky rivers/ponds or at sites just after large rainfall as the filter will soon clog and you will be unable to pass a sufficient volume of water through for analysis.
Try using an extendable pole or attaching the ladle to a long stick to get better access to the water. It is important to collect as best a representative sample as possible and avoid collecting the entire sample from a very small proportion of the site. Make sure you write down how much perimeter was accessible.
This could cause issues with the analysis (such as inhibition of the sample). Whilst in most cases our laboratory analysis procedures mean that we can account for this, we cannot guarantee reliable sample analysis if the sample is highly inhibited by such substances.
Where possible, try to avoid sampling sediment and highly untypical areas, make a note on the sample collection form of any abnormalities so we can use it in the interpretation of results.
Avoid collecting sediment. Where possible, samples should be collected where the pond/river is at least a minimum depth of 10cm to avoid sediment. If unavoidable, please contact us for further advice.
Streams and rivers: (less than 10m wide) less than 50cm deep. Where possible try to avoid entering the water system and collect samples from either side of the bank. If it is necessary and safe to enter the water system, collect samples following a zig-zag pattern as shown in the diagram below. Ensure that you sample in an upstream direction to avoid collection of sediment which may contain historical/ancient preserved DNA from the target species. If you enter the watercourse, ensure that you implement full biosecurity protocols to avoid the spread of disease such as crayfish plague and invasive non-native species.
Large rivers: For larger, more difficult to access rivers it may be necessary to use multiple collection kits in order to ensure reliability of species presence/absence. If you are unable to enter the water course as in the methods above then try where possible to collect samples from the edges, collecting a sub-sample every few metres in order to get a representative sample from the site.
If studying a large river system, it may be necessary to collect multiple samples at multiple points. For example, the collection of a sample every 500-1000m within a river. This is to ensure that you identify any populations which may be fragmented within the stream. Collecting multiple samples also enables the user to accurately indicate where a population may lie within the stream as opposed to only collecting one sample and only knowing if the species is present or absent.
For best results, return samples back to the lab as soon as possible. The longer the sample is stored before analysis, the more opportunity and time has passed which could lead to increased degradation of the DNA. Post sample collection, samples can be stored for a maximum of 2 weeks at room temperature and 4 weeks in the fridge. If you are working on a large project, feel free to store samples briefly until you have a larger batch to return to the lab.
Ideally next day delivery, using a courier of your choice. Try not to send on the weekend as we are unable to control the conditions in which the couriers store the kits, especially during hot weather.
All samples are analysed for the presence of target species eDNA following scientifically published eDNA assays and protocols which have been thoroughly tested and verified for use by SureScreen Scientifics.
The analysis is conducted in two phases. The sample first goes through an extraction process, where the filter is incubated in order to obtain any DNA within the sample. The extracted sample is then tested via real time PCR (also called qPCR) for each of the selected target species. This process uses species-specific molecular markers (known as primers) to amplify a select part of the DNA, allowing it to be detected and measured in ‘real time’ as the analytical process develops. qPCR combines amplification and detection of target DNA into a single step. With qPCR, fluorescent dyes specific to the target sequence are used to label targeted PCR products during thermal cycling. The accumulation of fluorescent signals during this reaction is measured for fast and objective data analysis. The primers used in this process are specific to a part of mitochondrial DNA only found in each individual species. Separate primers are used for each of the species, ensuring no DNA from any other species present in the water is amplified.
If target species DNA is present, the DNA is amplified up to a detectable level, resulting in positive species detection. If target species DNA is not present then amplification does not occur, and a negative result is recorded. If our internal control is not detected within the sample, then there is a possibility that the sample is inhibited. The sample is then diluted and re-analysed to test for inhibition.
All of our eDNA assays are reported as either positive, negative or inconclusive. Positive results are given a score out of 12, for how many of the 12 qPCR replicates were found to be positive. Whilst a high score such as 12/12 could appear to suggest a higher amount of DNA within the sample (when compared to one with a score of 1/12), it does not always mean a higher population at the survey site. This is due to an extremely high number of variable factors which can affect eDNA sampling and the distribution of eDNA across ecological sites. Such factors which can affect detection include differences in seasonal species-specific activity levels; how a species sheds DNA; size of site; population size; population locality to sample collection area; weather (including temperature and flow rate); and the presence of bacteria and inhibitory molecules etc. Without having an accurate picture of all of these factors it can be difficult to link DNA concentrations within the sample to population sizes within the sample site.
At the moment, these assays (with the exception of the GCN assay), without specific approval of legislators, cannot be used on legal basis to support planning and building applications. However, it can be a useful survey tool for the primary detection of populations as a complimentary survey technique.
The only eDNA species/method currently approved by Natural England is the one for GCN. All other species are not currently approved by any approval body; however, we are working towards this with the environment agency taking an assisting role with the validation of our method for white-clawed crayfish.
eDNA should not be used as a replacement for established/traditional survey methods. In its present form it is designed to be complimentary to these methods, as a primary survey technique which can enable the screening of large areas in a short amount of time with minimal training and licensing needs.
We have been conducting experiments to determine the rate at which the DNA degrades from the environment once a crayfish population is no longer present. Result indicate that the DNA levels drop below detectable amounts no later than one month (typically 7-14 days after the species have left the water), however, can last longer in sediment so be careful to not disturb it. This means that there is minimal chance of detecting a population of crayfish which have been absent for several months.
With invasive non-native species (INNS) and disease such as crayfish plague being a large problem for native species populations, it is highly important that biosecurity measures are followed when collecting eDNA samples. We have tried to design a biosecurity friendly eDNA kit which includes single use components and therefore reduces the risk of transferring disease/INNS from site to site. However, it is also important that the end-user thoroughly cleans any additional equipment, wellies, and clothes which they take to any site before moving onto a new site to reduce the risk of transferring any disease/INNS.