
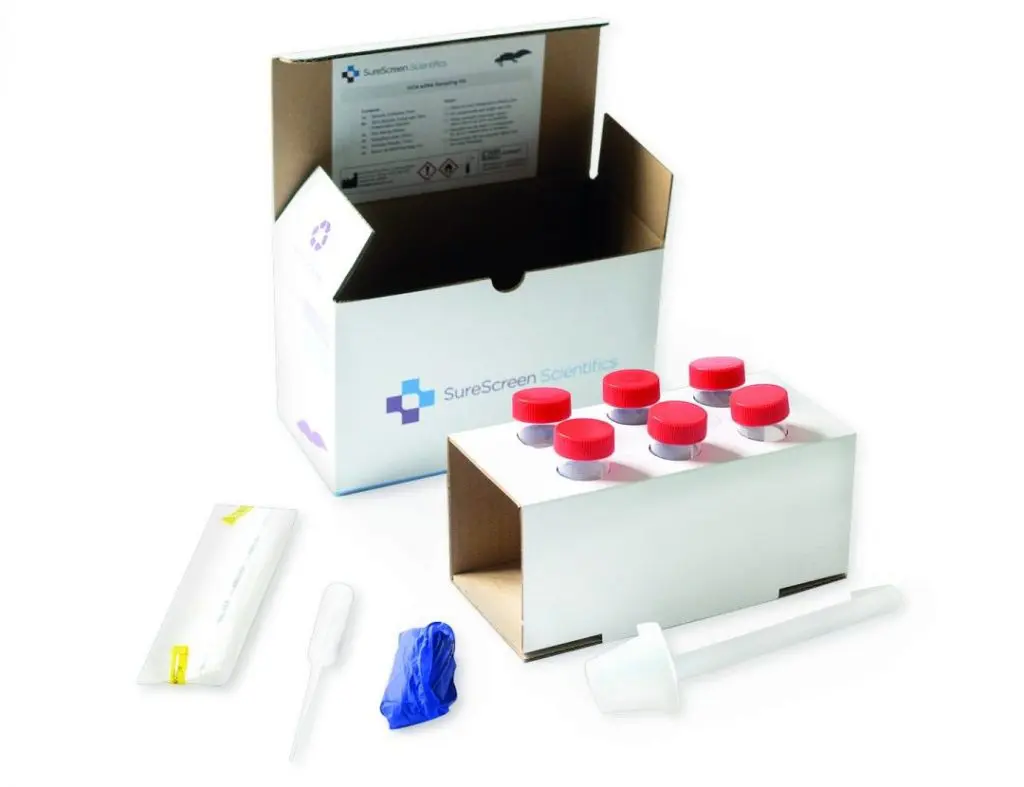
Great crested newt eDNA sample collection guide
- Articles
- Sample Collection Guidance


Step 1
Identify 20 evenly spaced sites around the pond perimeter- include areas likely to contain GCN.

Step 2
Put on the gloves provided. Gloves should be worn at all times during collection.

Step 3
Collect one ladle of pond water from each pre-identified sites. Gently mix the water column and avoid disturbing sediment.

Step 4
Transfer the ladle full of water to the bag provided. Avoid entering the water and ensure that any sediment in the pond is not disturbed.

Step 5
Repeat steps 4-5 at each of the sites identified in step 1 until 20 ladles of water have been placed within the bag.

Step 6
Once all sites have been sampled, tightly scrunch the bag and shake vigorously for 10 seconds (to mix any DNA within the sample equally).

Step 7
Using the pipette, transfer 15ml of water from the bag to each of the preservative filled tubes. You will need to use the pipette multiple times to take the level of the solution from 35ml to 50ml.
Repeat this step for all 6 tubes.

Step 8
Close and ensure the lids are tight/not cross threaded. Shake vigorously for 3 seconds. Leaky tubes = DNA loss and could cross contaminate other samples.

Step 9
Place all the tubes in the rack inside in the box. Fill in the form, place it in the box with the sample.

Step 10
Return to the laboratory for analysis.

Step 11
Results will be sent via the provided email address within your selected turnaround time.
Note: Mix the water column as you collect the sample, but avoid stirring or disturbing sediment. Avoid entering the water where possible and ensure that any sediment in the pond is not disturbed. Disturbed sediment could cause inhibition of the sample analysis, or disturb ancient DNA which has been preserved within the sediment.
Note: the tube is prefilled to 35ml, adding 15ml will require several pipette movements from the bag to the tube. See markings on tube for 50ml mark.
Note: Cross threaded/leaky tubes result in loss of DNA and could cross contaminate other samples within your order. A significant loss of liquid before analysis could result in an inconclusive result.
Note: you can return unwanted components back to the laboratory for recycling/disposal.


Kits are guaranteed to last for the entire season which is in line with Natural England guidelines (all kits guaranteed until 30th June minimum or for 3 months – whichever is the latest date). However, if they are kept in good conditions (cool, dark) the kits should last longer than that, but outside the use by date Natural England will not accept the result.
One kit is required for one pond up to approximately 1 hectare (2.5 acres). For ponds larger than this it is recommended to collect one kit per hectare. Generally, the most accepted way of collecting samples for a large pond of say 2 hectares, is that two kits will be used, one to collect 20 sub-samples around one half of the pond and the other kit to collect 20 sub-samples around the other half of the pond. These kits are analysed separately (and therefore charged as two kits and two analyses) as to not dilute the sample and give best chance to detect GCN DNA.
Yes, you need separate kit for each pond.
Natural England has defined the GCN eDNA sample collection period to coincide with great crested newt breeding and will accept only samples collected between the 15th of April to 30th June. Samples may be taken outside of this time, provided GCN activity is noted in the area (positive result). Dates outside of this can sometimes be accepted on a case-by-case basis with prior Natural England approval.
eDNA samples can be collected at any time of day and in any reasonable weather conditions, including light rain. It may be best to avoid heavy rain as this makes sampling more difficult and might increase the risk of cross contamination (e.g. splashing of mud which could contain great crested newt DNA from wet ground). There is evidence that unpreserved amphibian eDNA decays slightly more quickly in full sun than shaded conditions, becoming undetectable after 8 and 11 days respectively, but as long as samples are preserved the impact on detection should be slight. On the other hand, we advise not to sample during or recently after heavy rainfall – the DNA is more diluted than normal.
Existing data shows that eDNA can be very patchy depending on where the animals have been. By sampling in many areas, you considerably increase your chance of collecting their DNA successfully.
DNA ‘sinks’ and so will often be present in larger amounts close to the pond bottom. However, it is important not to collect sediment because DNA may persist in the sediment for substantially longer than in the water column. If you collect sediment, there is a risk your sample might show a false positive indicating great crested newts were present recently, when in fact this was a long time in the past.
Try using an extendable pole or attaching the ladle to a long stick to get better access to the water. It is important to collect as best a representative sample as possible and avoid collecting the entire sample from a very small proportion of the site. Make sure you write down how much perimeter was accessible.
This can cause issues with analysis (such as inhibition of the sample). Whilst in most cases our laboratory procedures mean that we can account for this, we cannot guarantee reliable analysis if the sample is highly inhibited. Where possible, try to avoid sediment and highly untypical areas, make a note on the sample collection form of any abnormalities so we can use it in the interpretation of results. If these conditions occur after rain or a heavy storm, try to collect sample on different day, 3-5 days later, when the sediment has settled and the risk of picking up in the sample is reduced.
Avoid collecting sediment. Where possible, samples should be collected where the pond/river is at least a minimum depth of 10cm to avoid sediment. If unavoidable, please contact us for further advice.
For best results, return samples back to the lab as soon as possible. The longer the sample is stored before analysis, the more opportunity and time has passed which could lead to increased degradation of the GCN DNA by temperature, light, enzymes etc. Post sample collection, samples can be stored for a maximum of 2 weeks at room temperature and 4 weeks in the fridge. If you are working on a large project, feel free to store samples briefly until you have a larger batch to return to the lab.
Ideally next day delivery, using a courier of your choice, try not to send on weekend as we are unable to control the conditions in which the couriers store the kits, especially during hot weather.
Please send completed kits back to our laboratory by courier at: SureScreen Scientifics Ltd, Morley Retreat, Church Lane, Morley, Derbyshire, DE7 6DE. We also accept hand-delivered kits between 08.00 and 16.30 Monday to Friday (16:00 on Friday), and by appointment 24/7. For a collection charge of £40 per batch, we can arrange sample collection from you with our preferred courier, DPD. However, it may be logistically or financially beneficial to arrange the return courier service yourself.
The ethanol-based preservative in the sample bottle will slow, but not eliminate, degradation of DNA. Keeping the samples refrigerated also slows this process. As long as your kits arrive at our labs within three weeks of collection the analysis/results are unlikely to be affected.
At SureScreen Scientifics GCN eDNA samples are analysed for the presence of GCN eDNA following the protocol stated in DEFRA WC1067 ‘Analytical and methodological development for improved surveillance of the Great Crested Newt, Appendix 5.’ (Biggs et al. 2014). Each of the 6 sub-sample tubes are first centrifuged and pooled together into a single sample which then undergoes DNA extraction. The extracted sample is then analysed using real time PCR (qPCR), which uses species-specific molecular markers to amplify GCN DNA within a sample. These markers are unique to GCN DNA, meaning that there should be no detection of closely related species. If GCN DNA is present, the DNA is amplified up to a detectable level, resulting in positive species detection. If GCN DNA is not present then amplification does not occur, and a negative result is recorded. Analysis of eDNA requires scrupulous attention to detail to prevent risk of contamination. True positive controls, negative controls and spiked synthetic DNA are included in every analysis and have to be correct before a result is reported. Stages of the DNA analysis are also conducted in different buildings at our premises for added security.
Lab analysis detecting GCN DNA is highly sensitive. A study from 2014 focused on GCN shows that eDNA analysis detected GCN in 99.3 % (139 times out of 140 samples from ponds where we knew newts were present). There were no false positive results.
Once your results are available, they will be sent to the email address provided on the sample collection form before the deadline. If you have not received your results before this date, please get in touch and we will assist you (please make sure that you check your spam or junk email folder).
A positive result is indicative of GCN presence at the time of sampling or within the 3-4 weeks (GCN eDNA degrades to below detectable levels between 7-21 days). A negative result suggests that there are no great crested newts within the sample area. Inconclusive results are rare, however, indicates that although GCN DNA has not been detected there may be some underlying degradation or inhibition which could be affecting the interpretation of results. This could be due to sediment, algae or plant matter within the sample, or an unusually high concentration of inhibitory molecules within the pond water. For inconclusive results, it is recommended that analysis is repeated with a fresh sample from the site in question.
We have a number of checks on the sample to ensure quality results are provided, these are: quality check, degradation check and inhibition check.
SIC: Sample Integrity Check [Pass/Fail]
When samples are received in the laboratory, they are inspected for any tube leakage, suitability of sample (not too much mud or weed etc.) and absence of any factors that could potentially lead to inconclusive results.
DC: Degradation Check [Pass/Fail]
Analysis of the spiked DNA marker to see if there has been degradation of the kit or sample between the date it was made to the date of analysis. Degradation of the spiked DNA marker may indicate a risk of false negative results.
IC: Inhibition Check [Pass/Fail]
The presence of inhibitors within a sample are assessed using a DNA marker. If inhibition is detected, samples are purified and re-analysed. Inhibitors cannot always be removed, if the inhibition check fails, the sample should be re-collected.
Our eDNA assays are reported as either positive, negative or inconclusive. Positive results are given a score out of 12, for how many of the 12 qPCR replicates were found to be positive. Whilst a high score such as 12/12 could appear to suggest a higher amount of DNA within the sample when compared to one with a score of 1/12, it does not always mean a higher population at the survey site. This is due to an extremely high number of variable factors which can affect eDNA sampling and the distribution of eDNA across sites. Such factors which can affect detection include differences in seasonal species-specific activity levels; how a species sheds DNA; size of site; population size; population locality to sample collection area; weather (including temperature and flow rate); and the presence of bacteria and inhibitory molecules etc. Without having an accurate picture of all of these factors it can be difficult to link DNA concentrations within the sample to population sizes within the sample site.
Like any survey method, eDNA is not fool proof and does have its limitations. The result you obtain is only as good as the sample collection, eDNA distribution in a pond is not homogeneous, so if subsamples were not taken effectively, we may not be able to detect eDNA. High levels of inhibition or degradation of sample between sampling and analysis can also result in false negative results.
False positive detections can occur from time to time with eDNA sampling. The important thing here is to have as much information about the site as possible to identify if it could be a false positive and interpret the meaning of this result. Often false positives can occur via contamination (at any stage during the collection and analysis process) or cross-contamination of the site (eDNA spread between ponds via birds, dogs, water inflow, recreational activities etc.), prior to sample collection. Due to the sensitivity of eDNA testing, it is also possible that GCN are present within the pond at extremely low levels, below detection of traditional methods. If your sample is reported as positive and you expect this to be a false positive then we would recommend re-testing, with additional samples for security.
Inhibition can be caused by a number of factors. The presence of chemical run off from farms/industry/fertilisers etc is one such cause which is widely reported to affect the analytical process (namely the extraction and qPCR steps). Inhibition happens when a substance or molecule is present in a sample which prevents the PCR process from occurring efficiently. We are able to detect for this by including a known quantity of non-target DNA (we call this DNA spike) within each kit. When we receive kits back into the laboratory, we screen them for both GCN and this DNA spike. If we fail to detect this DNA spike, it can be hypothesised that the sample is subjected to some form of inhibition, which affects the amplification process of the qPCR. In some cases, the inhibition can be overcome by diluting the sample and re-analysing it. Usually this resolves about 60% of inhibited samples and results in a reliable result. However, if your sample was reported as inconclusive, inhibition was still present as the spike DNA was not found to be present even after diluting. Further dilutions and tests reported the same conclusion. For inconclusive results, it is recommended that analysis is repeated with a fresh sample from the site in question.
In ponds, when collecting subsamples make sure not to disturb sediment which may contain historical DNA from the target species (in some cases can be present for up to 2 years post species removal form the pond). If needed use an extendable pole to prevent such disturbances.
SureScreen Scientifics participates in the proficiency testing, have done since its birth in 2017 and were actively involved in setting the scheme up. Our results will be accepted by Natural England this season. We are proud to say that we scored 100% in the latest proficiency test.
The purchase order /reference number is required to be on the form to help smooth out the payment process. It is mainly serving the purpose of being the reference on the invoice that you are sent regarding the charges for analysis, so it is easy to identify what project the charges relate to. This should be generated by yourselves – if you don’t use purchase orders, just put a reference in there such as your name or project name. Feel free to use the same purchase order for all forms. If you have paid upfront for the analysis, please state your payment reference number. We are unable to begin processing a sample without a purchase order provided on the sample collection form. This means that there might be a delay to sample processing.
We are unable to analyse kits manufactured by other eDNA service providers as we would not be able to detect their DNA spike to check for degradation.
For Natural England to accept results, these should be used within 3 months of manufacture. This applies to most cases. If you have expired kits – do not dispose of them, make use of old/out of date kits by using our GCN eDNA Refill Kit which allows you to replace the critical parts in an old kit and use it as brand new – reducing excess, unnecessary waste. If it is not critical for approval by Natural England (I.e. for school project/conservation) and only when not planning and building related, then older kits can be used. Due to the nature of the DNA spike, which is contained within them, the kits typically have a use by date of 3 to 6 months from manufacture for which the stability of the DNA spike is guaranteed. The DNA spike is important in enabling the detection of any inhibition of the sample or degradation of sample post collection in the case that a sample turns out to be negative. With the older kits we are unable to guarantee that DNA spike will still be present as it has most likely degraded. Therefore, should the sample be negative for both GCN DNA and DNA spike, we would have to report the sample as inconclusive.
Instead of disposing of your unused kit, you can buy our GCN eDNA Refill Kit and use it as brand new.
Otherwise, kit components can be disposed of or recycled according to your locally available recycling facilities. Sample preservative solution can be disposed of by pouring down the sink with copious amounts of water. If you would prefer, you can return kits to us for correct disposal.
The preservative is an ethanol/sodium acetate buffer solution containing 95% absolute ethanol and 5% sodium acetate solution at pH 5.2. We can’t provide a COSHH sheet as we have one just for preparing DNA preservative, not for using in field. Get in touch for more information.
We have found over the years that on average 88% of kits come back to us. The rest don’t come back to us for several reasons such as ponds being dried up. However, if this happens to you, then it will have only cost you the price of the kit – in this case, the analysis is not charged.
Yes, we can ship to these locations for an additional delivery fee. Contact us for more details.
For queries of this nature we would advise you to contact Natural England or the relevant local authority for further advice.